Predictive coding and active sensing

Predictive coding has been proposed as a method to create efficient neural codes. It postulates that the brain learns and represents information about the world through hierarchical interactions across cortical areas and layers. Active sensing, on the other hand, is a strategy employed by animals to optimally sample relevant sensory information to effectively update the internal representation. Together, predictive coding and active sensing form a biologically plausible model of efficient and robust perception and action.
Our goal is to understand the neural correlates of predictive coding and active sensing. To achieve this, we incorporate data-driven connectivity, cell types, and neural dynamics to construct predictive coding and active sensing models of cortical circuits and other neural systems.
Related publications (selected):
2025, A. Sharafeldin*, H. Choi*, “Cell-type-specific encoding of prediction and reward in cortical microcircuits during novelty detection”, bioRxiv preprint: doi.org/10.1101/2025.05.13.653877
2024, A. H. Balwani*, S. Cho, H. Choi*, “Exploring the architectural biases of the canonical cortical microcircuit”, Accepted, Neural Computation
2024, A. Sharafeldin*, N. Imam*, H. Choi*, “Active sensing with predictive coding and uncertainty minimization”, Patterns 5: 100983
2023, D.G. Wyrick, N. Cain, R.S. Larsen, J. Lecoq, M. Valley, R. Ahmed, J. Bowlus, G. Boyer, S. Caldejon, L. Casal, M. Chvilicek, M. DePartee, P.A.Groblewski, C. Huang, K. Johnson, I. Kato, J. Larkin, E. Lee, E, Liang, J. Luviano, K. Mace, C. Nayan, T. Nguyen, M. Reding, S. Seid, J. Sevigny, M. Stoecklin, A. Williford, H. Choi*+, M. Garrett*+, L. Mazzucato*+, “Differential encoding of temporal context and expectation under representational drift across hierarchically connected areas” bioRxiv preprint: doi.org/10.1101/2023.06.02.543483
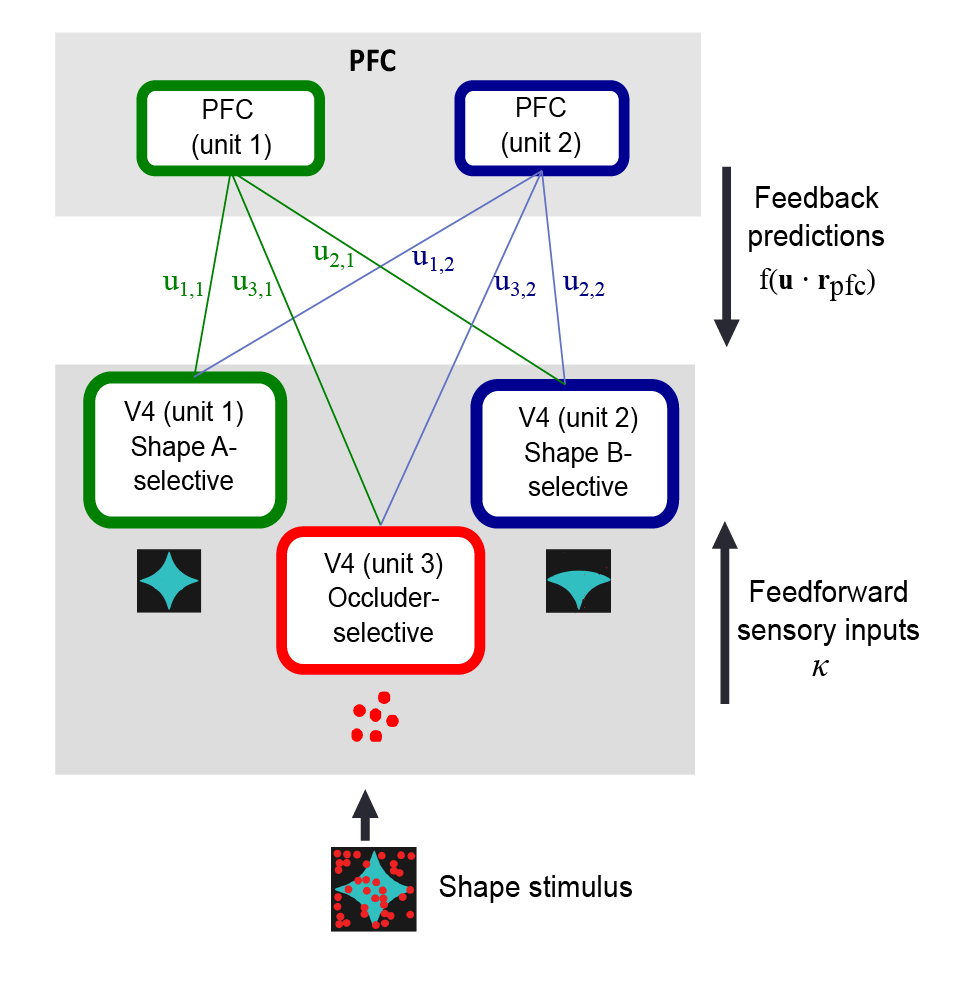
2018, H. Choi*, A. Pasupathy, E. Shea-Brown, “Predictive coding in area V4: dynamical shape discrimination under partial occlusion”, Neural Computation 30(5):1209–1257
2017, A.M. Fyall+, Y. El-Shamayleh+, H. Choi, E. Shea-Brown, A. Pasupathy, “Dynamic representation of partially occluded objects in primate prefrontal and visual cortex”, eLife 6:e25784
Related presentations (selected):
2023, A. Balwani, S. Cho, H. Choi, “Exploring the architectural biases of the canonical cortical microcircuit” Cosyne, Montreal, Canada. Watch Aish’s talk here!
Topology and computations of anatomical and functional neural networks


The exquisite ability of the brain to perform adaptable, efficient, and robust computation in changing environments is attributed to complex connectivity structures of underlying networks. Across multiple scales ranging from microscopic cell-to-cell connectivity to a large-scale network of brain regions, biological neural networks are characterized by their topological complexities. The distinctive topological motifs of the anatomical connectivity affect network dynamics and neural coding. On the other hand, functional connectivity constructed from correlated neural activities, is shaped by the underlying anatomical network structures as well as stimulus complexity and behavioral states. Functional networks inferred from population activities, furthermore, reflect information transmissions among neurons, neuronal populations, and brain regions.
We use data-driven modeling to probe the precise connections between topology and computation in both functional and anatomical networks of neurons, in close collaboration with experimental neuroscientists.
Related publications (selected):
2025, A. Balwani*, A. Q. Wang, F. Najafi, H. Choi*, “Constructing biologically constrained RNNs via Dale’s backprop and topologically-informed pruning“, bioRxiv preprint: doi.org/10.1101/2025.01.09.632231
2024, D. Tang*, J. Zylberberg*+, X. Jia*+, H. Choi*+, “Stimulus type shapes the topology of cellular functional networks in mouse visual cortex”, Nature Communications 15: 5753
2024, C. Li, S. H. Kim, C. Rodgers, H. Choi, A. Wu*, “One-hot generalized linear model for switching brain state discovery”, The 12th International Conference on Learning Representations (ICLR): 1-22
2023, U.B. Sikandar*, H. Choi, J. Putney, H. Yang, S. Ferrari, S. Sponberg, “Predicting visually modulated precisely-timed spikes across a coordinated and comprehensive motor program”, International Joint Conference on Neural Networks (IJCNN) :1–8
2020, J.H. Siegle+, X. Jia+, S. Durand, S. Gale, C. Bennett, N. Graddis, G. Heller, T. Ramirez, H. Choi, J.A. Luviano, …, S.R. Olsen, C. Koch, “Survey of spiking in the mouse visual system reveals functional hierarchy”, Nature 592:86–92
2019, J.A. Harris+, S. Mihalas+, K.E. Hirokawa, J.D. Whitesell, H. Choi, …, C. Koch, H. Zeng, “Hierarchical organization of cortical and thalamic connectivity”, Nature 575:195-202
Neural dynamics and coding of single cells, populations, and networks


Neurons communicate diverse information of sensory stimuli, decision signals, and motor commands, using their rich dynamics at both single cell and population levels. Their operating modes are characterized by different spiking patterns. These patterns are determined by various biological factors: intrinsic properties of individual neurons such as ion channels and synaptic variables, and network properties defining population composition and connectivity of spatially-structured heterogeneous neurons. In addition, sensory stimuli, internal states, and behavioral tasks shape neural dynamics at both the local and global scales.
We investigate how changes in local (intrinsic) and global (network) variables instigate transitions across different phases of single-neuron spiking dynamics as well as population dynamics described by excitatory-inhibitory balance, synchronizability, sensitivity, and dimensionality.
Related publications (selected):
2025, S.H. Kim*, H. Choi*, “Inhibitory cell type heterogeneity in a spatially structured mean-field model of V1“, bioRxiv preprint: doi.org/10.1101/2025.03.13.643046
2025, Z. Mobille*, U.B. Sikandar, S. Sponberg, H. Choi*, “Temporal resolution of spike coding in feedforward networks with signal convergence and divergence”, PLOS Computational Biology 21(4): e1012971
2025, J. Del Rosario, S. Coletta+, S. H. Kim+, Z. Mobille, K. Peelman, B. Williams, A. J. Otsuki, A. Del Castillo Valerio, K. Worden, L. T. Blanpain, L. Lovell, H. Choi, B. Haider*, “Lateral inhibition in V1 controls neural and perceptual contrast sensitivity“, Nature Neuroscience 28: 836–847
2019, H. Choi*, S. Mihalas, “Synchronization dependent on spatial structures of a mesoscopic whole-brain network”, PLOS Computational Biology 15(4): e1006978
2014, H. Choi, L. Zhang, M.S. Cembrowski, C.F. Sabottke, A.L. Markowitz, D.A. Butts, W.L. Kath, J.H. Singer*, and H. Riecke*, “Intrinsic bursting of AII amacrine cells underlies oscillations in rd1 mouse retina”, Journal of Neurophysiology 112: 1491-1504